In our previous post we decided that we will create a bioreactor that is able to compress 3D tissue cultures and integrate into a high throughput workflow, and discussed the novelty and potential applications of such a device.
Here, we discuss some initial ideas and some early phase decisions that came up in our design process.
Microtiter plates, sometimes called microwell plates, have become a standard tool for analytical research in the life sciences. They consist of a flat plastic plate with wells arranged in a 2:3 ratio, and standard plate formats include 6-, 24-, 96-, and 384- well matrices. Sigma-Aldrich, a popular lab equipment retailer, has hundreds of products from dozens of brands available online in this format. Most tools for high-throughput life sciences workflows interoperate with this format, so it seemed like a natural choice to center our bioreactor around a microtiter plate.

Lab equipment from the ISMMS Integrated Screening Core. At bottom left is a plate sealer with a microtiter plate in its tray. Also pictured are a plate storage unit at top, and a plate handling robot at center.
An early decision was which well format we would target. The tradeoff is between the complexity of the bioreactor in handling more wells at once, and the overall throughput of one device. There was some discussion about what constitutes “high throughput” in the cartilage and tissue engineering field, with some students arguing that dozens of samples would be a significant improvement over current published methods for a compression bioreactor, while others insisted that we could shoot for thousands of samples and only at this scale would we achieve something “high-throughput”.
To minimize the complexity incurred by this tradeoff, we decided that we should try to delegate as many tasks as possible to existing devices in the high throughput workflow. There are already complicated but well-supported solutions for moving plate-size objects around and dispensing liquids into them. It would be costly and redundant for us to implement such functions in our own bioreactor compared with our design facilitating other devices coming in to do the same work.
We would then, however, need to design our bioreactor around the requirements of those devices. Regardless of the choice of well format, it was pretty clear our bioreactor would need a “nest”, which is where the microtiter plate would sit. Several nests and a Tecan plate manipulator are shown below.
Different plate handling robots have different capabilities for manipulating the plates; for example, both of the previously shown robots can insert a plate into a shelf since they hold the plate from the side. Other robots grip the plate from above, however, and with this configuration our device must expose the entire top surface of the plate in order for it to be removed.
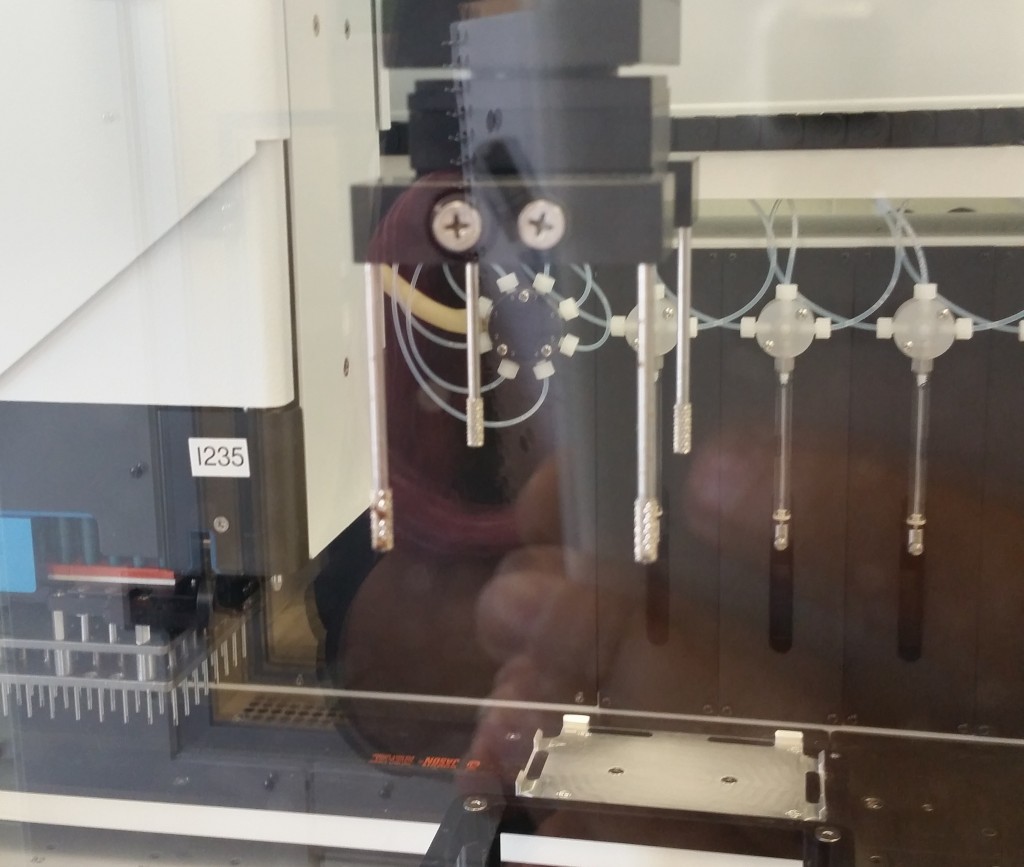
This plate handler grips the plate from the top, using the four grippy rods, and therefore cannot insert the plate into a shelf-like space—it can only drop it into an exposed nest.
We were informed that the latter restriction is pretty common, and the Tecan equipment we would have access to would likely have this same restriction, so we decided to assume a top-gripping plate handler going forward. We would want a robot to be able to manipulate the plate into and out of our device, so that regardless of the well format, a plate handler could swap in dozens of plates for compression in sequence. This is what allows us to jump from, e.g., 96 samples to thousands of samples, if a robot can automate the application of our bioreactor’s compression function to many 96-well plates over the course of one experiment.
Assuming a top-gripping robot and that our compression action would also be applied from the top of a standard microtiter plate required us to consider how our device could alternately attach to the wells from above and subsequently allow them to be exposed for manipulation and liquid exchange. There are a few approaches used by existing devices, such as a “sliding drawer” approach and the “panini press” approach pictured below.

Two approaches to exposing microwell plates for manipulation; left: “sliding drawer” approach as used by a plate reader, right: “panini press” approach as used by a thermocycler
Both of these approaches require a large moving part: either a drawer or a hinge, which would have to be actuated separately from the compression action applied to the wells. In the case of the hinge, a lot of the device would have to live inside a heavy “lid” that might require a significant amount of force to lift off or lock down onto the the plate.
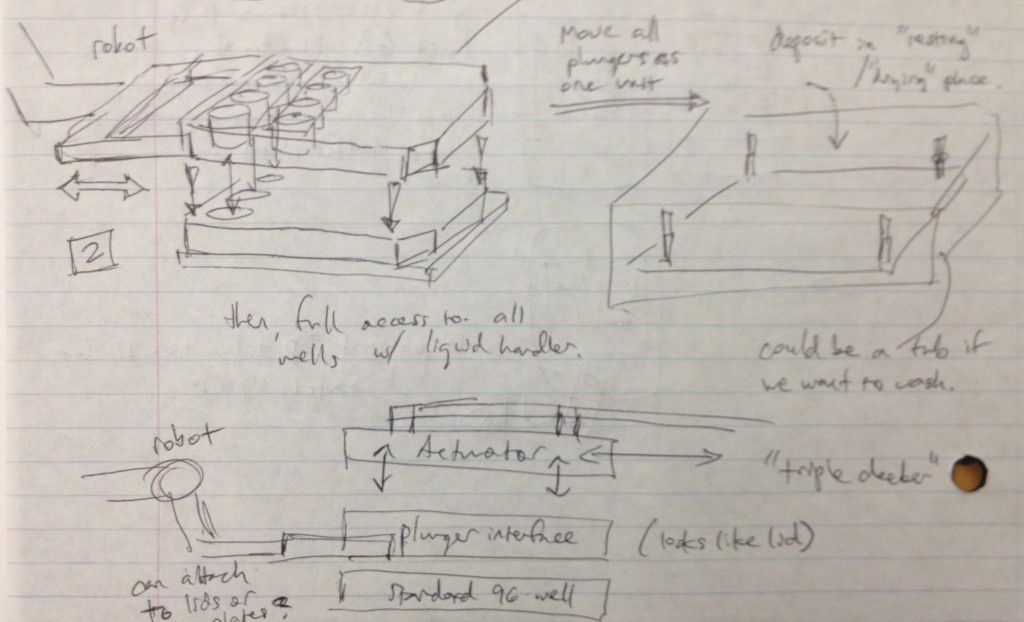
It occurred to us at this point that in order to maintain sterility between samples in wells from different plates, there would need to be an “adapter lid” that can apply the compression to the tissue sample while avoiding direct contact between the culture and the actuation mechanism. This way, one actuation mechanism can operate on multiple adapter lid + microtiter plate pairings, manipulated by the robot, to enable the thousand-sample scale described previously. If the adapter lid is a plastic part, it could be cheaply printed for many plates and autoclaved between runs, and if it were roughly the size of a microtiter plate itself, the plate manipulator robot could lift the adapter lid off of the plate and place it separately onto another nest to allow access to the plate’s wells by a liquid handler. These ideas are expressed in the following design sketches.
Dan Felsenfeld of the ISMMS Integrated Screening Core raised the point that if our device lived mostly underneath the plate, like a microplate shaker, it could latch onto it and actuate the plungers from below. This saves horizontal space, which is at a premium inside a liquid handler, and could allow for actuation and latching to be driven by one motor. This idea is incorporated into the following sketch.
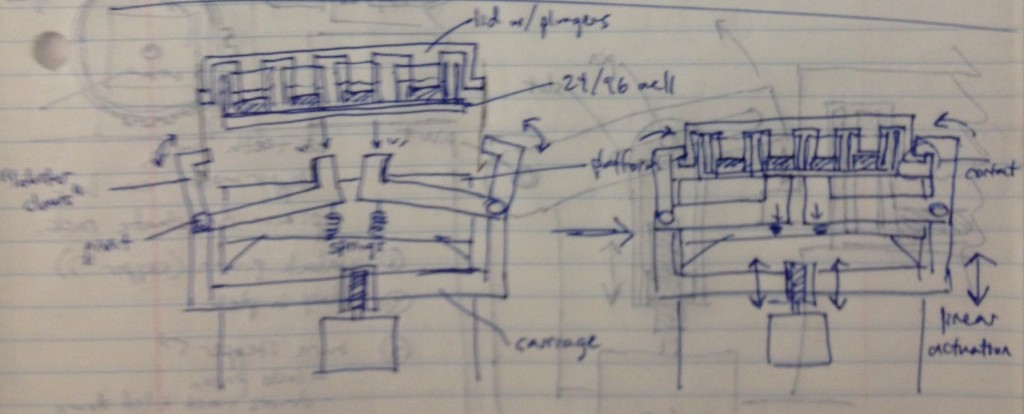
In this design, dropping the plate onto the device causes the rotating claws to latch onto the lid from the sides. The claws can then be actuated by a stepper motor and screw drive from below.
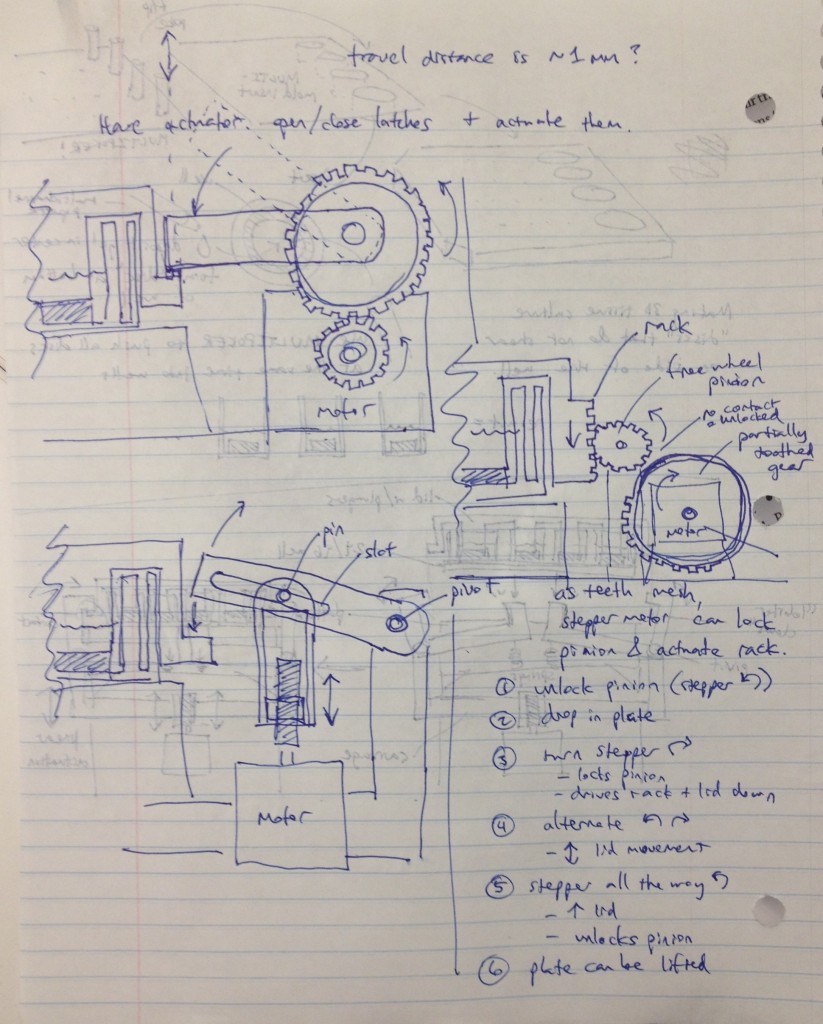
We recognized that this particular design, while attractively compact and utilizing only one motor for actuation, requires a fair number of moving parts. Some of these moving parts may have tight tolerances, particularly the arms that must latch neatly over the edge of the lids while releasing the plate promptly when it is lifted. We considered whether we could move more of the mechanism off to the sides and reduce the number of tricky parts.
Three possible simpler designs are shown in the sketch above. The rightmost sketch involves a rack that would be printed on the edge of the adapter lid, which engages a freewheel pinion as the plate is dropped onto the bioreactor’s nest. A partially toothed gear could then be rotated by a stepper motor to lock the lid via the freewheel pinion, and then depress it, applying force to the tissue samples.
The other two designs to the left top and left bottom are most analogous to a traffic gate:
and involve the use of a stepper motor or linear actuator to move an arm up and down that depresses the adapter lid. By rotating the arm sufficiently upward, it can be moved out of the way of any plate manipulators that require access from above.
To keep the motion of these designs symmetrical, the mechanism could be duplicated onto the other side of the device and the arms and motors would be moved in unison via software control.
The bottom-left design was considered most attractive overall due to its simplicity and the possibility of altering the compression force by changing the thread count of the screw. Cheap linear actuators based off an internal screw drive are readily available from Firgelli and they are also apparently controllable by Arduino boards.
Stay tuned for a post detailing how we chose our plate format (24-well vs. 96-well?) and the loading functionality of the bioreactor (hydrostatic pressure vs unconfined compression?) along with other important design decisions.